Molecular Basis of Inheritance Class 12 | Chapter 6 | Biology | CBSE |
Table of Contents
Molecular basis of Inheritance Class 12 | Chapter 6 | Biology | CBSE
Experiment to prove DNA as a Genetic material
Griffith Experiment (Transforming Principle)
Griffith was studying pathogenicity of different strains of bacterium Streptococcus pneumoniae which is also known as Diplococcus or Pneumococcus.
This bacterium has two strains – virulent and non virulent.
Virulent – S type :
- This strain causes pneumonia.
- This is known as S type because when grown on suitable medium they form smooth colonies.
- These diplococci are covered by a sheath of mucilage (polysaccharide) around them.
- The sheath is not only the cause of toxigenicity but also protects the bacteria from phagocytes of the host.
Non virulent – R type :
It do not produce the disease.
They form irregular or rough colonies.
These diplococci are devoid of mucilage sheath.
- Griffith tested the virulence of these two strains by injecting the live R type and live S type bacteria separately into mice.
S strain ⇒ Inject into mice ⇒ Mice die
R strain ⇒ Inject into mice ⇒ Mice live
He found that R type bacteria did not produce any disease while the S type bacteria caused pneumonia and then lead to death of the mice.
By heating them, he was able to kill the bacteria. He observed that when heat – killed S strain bacteria injected into mice then it did not kill the mice.
S strain (heat killed) ⇒ Inject into mice ⇒ Mice live
Finally, he injected a combination of live R type and heat killed S type bacteria into mice which leads the death of mice.
S strain (heat killed) + R strain (live) ⇒ Inject into mice ⇒ Mice die.
- Moreover, he recovered living S bacteria from the dead mice.
Transforming Principle – The process of uptake of genetic material from the surrounding and conversion of 1 form of bacteria into the another form.
Biochemical characteristics of Transforming principle
Before the work of Avery, MacLeod and McCarty, the genetic material was thought to be protein.
But after their discovery, it was concluded that genetic material is DNA.
The above experiment reveals that DNA is responsible for transformation.
Transformation can also be observed in bacteria.
Example – Hemophilia influenza, Bacillus subtilis, Escherichia coli
Proof of DNA as a Genetic material
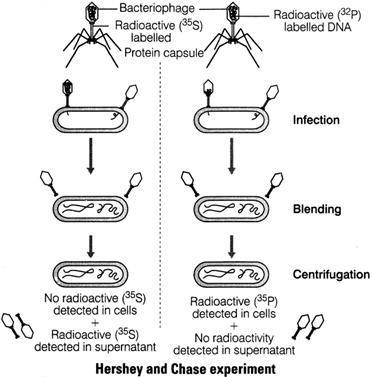
Hershey and Chase experiment
They took T2 bacteriophage which affect E. Coli.
T2 bacteriophage is made up of DNA and protein.
According to them only DNA of phage enter in bacterial cell and capsid remain outside the E – Coli.
Key feature of their discovery is that DNA contains phosphorus and protein contain sulphur.
| BATCH 1 | BATCH 2 |
| Phages were grown in culture having radioactive sulphur.
↓ |
Phages were grown in culture having radioactive phosphorus.
↓ |
| Sulphur mixed with protein.
↓ |
Phosphorus mixed with DNA.
↓ |
| Phages were allowed to infect E. coli.
↓ |
Phages were allowed to infect E.coli.
↓ |
| Now mixture is blended and centrifuged. | Now mixture is blended and centrifuged. |
- Phage DNA enter bacterial cell, but phage protein do not. Hence, they concluded that DNA is a genetic material not protein.
Central Dogma – Given by Crick
Central dogma defines, how DNA express in the cell.
Reverse Central Dogma
It occur in Retrovirus or RNA virus.
DNA Replication
It is the process by which DNA make its identical copy.
Property of DNA replication :
- It is always semi – conservative. It was given by Watson and Crick.
- It is semi – continuous.
Replication
- Ori (Origin of replication) – These are the specific sequence present within the DNA from where DNA replication get started.
- Unwindase/Helicase – It helps in unwinding of DNA double helical structure. It recognise origin of replication, get bind over it and start breaking hydrogen bond between double strand DNA.
- Replication fort – It is Y – shape structure where replication takes place.
- SSBP (Single strand binding protein) – They get bind over the single strand DNA and stabilise the structure.
- Topoisomerase – It remove the excess of stress produce during unwinding of DNA. It is also known as gyrase in case of prokaryotic cell.
- RNA polymerase/Primase – It synthesize a small RNA strand on the 3’ of the Template strand.
- DNA polymerase – It is the main polymerizing enzyme that add dNTPs (deoxynucleotide triphosphate) by following the complementary rule and synthesize DNA on the Template strand.
Dual role of dNTPs :
- Act as substrate.
- Provide energy for the polymerization.
Properties of DNA polymerase :
- It synthesize DNA.
- Help in removal of RNA primer.
- It has proof reading property – i.e. before adding a new base pair, it go back and check whether it add correct BP or not, if wrong BP added by mistake then it remove and add the correct BP.
Why DNA replication is semi – continuous ?
Because it take place in fragments as DNA is very long in size and required large amount of energy to become single strand and such huge amount of energy is not available in the cell.
Transcription
The process of formation of RNA on the DNA.
Transcription unit – The segment of DNA which undergo transcription.
Component of Transcription unit
Promoter – These are the specific sequence present on the DNA and that help in initiation of process of transcription.
They are present upstream to the structural gene.
They are located on the 5’ of the sense strand/coding strand or 3’ of the template strand of structural gene.
Terminator – These are the specific sequence present on the DNA and that help in completion of process of transcription.
They are present downstream to the structural gene.
They are located on the 3’ of the template strand or 5’ of the coding strand of structural gene.
It is made up of repeated sequence of adenine and thymine.
Structural gene
It is the part of the transcription unit, which get transcript in the form of RNA.
Structural gene have two type of sequence :
- Intron – These are the non – coding sequence
- Exon – These are the coding sequence
On the basis of type of sequence, structural gene is of two types :
- Split gene –
Intron sequence present.
Undergo splicing.
Present in eukaryotic cell.
- Non – split gene –
Intron sequence absent.
Do not undergo splicing.
Present in prokaryotic cell.
Types of structural gene –
| Polycistronic gene | Monocistronic gene |
| Here, gene transcribes RNA which codes for more than 1 protein. | Here, gene transcribes RNA which codes for only 1 protein. |
| It is seen in prokaryotes. | It is seen in eukaryotes. |
Process of Transcription
The steps for the transcription are:
- Initiation
- Elongation
- Termination
Initiation
RNA polymerase joins at promoter region. This joining is assessed by σ – factor.
Initiation of Transcription involve winding of RNA polymerase as Holoenzyme.
Elongation
Once promoter region has been recognise by RNA – polymerase as Holoenzyme, RNA – polymerase synthesize RNA over DNA.
Sigma – factor must be released from RNA – polymerase for elongation step.
Substrate during transcription are ribonucleotide triphosphate.
Termination
The process of elongation of RNA chain continue until terminator region is achieved.
Terminator region is G ≡ C rich region.
ρ – factor is required for termination process because it has ATP dependent RNA – DNA helicase activity. Thus, break DNA – RNA hybrid.
Maximum functions of enzymes are performed by RNA – polymerase.
Post Transcriptional changes
Splicing – It is the process of removing of intron and joining of adjacent exon.
During the splicing hnRNA is converted into functional mRNA.
The process of splicing takes place within the nucleus.
Splicing takes place with the help of SnRNPs (small nuclear ribonucleo protein).
Spliceosome – SnRNP get bind over the intron, to cause its splicing, then the structure that form after the binding of SnRNP over the intron is known as Spliceosome.
During the splicing, two transesterification reaction takes place.
Capping
Addition of unusual base pair i.e. 7 – methyl guanosine triphosphate at the 5’ of the mRNA is known as Capping.
It help in the initiation of process of Translation as during the translation ribosome recognise the 5’ of the mRNA due to the presence of this unusual base pair.
Tailing
Addition of poly adrenaline chain at the 3rd end of the mRNA.
It protect the mRNA from the Ribo nuclease enzyme present in the cytoplasm.
Transcription and Translation can be coupled in prokaryotic cell because
- Site of transcription and translation is same i.e. cytoplasm, as no nuclear membrane present around the nucleoid.
- In prokaryotic cell, intron is absent. So, RNA do not undergo splicing.
Why both strand of DNA do not get transcript ?
- If transcription occur on both the strand of the DNA, it lead to formation of complementary RNA strand, that get join and form the double strand DNA.
- If both the strand get transcript, then it lead to formation of 2 different type of RNA that form 2 different protein. If it happen, then genetic information get complicated.
Transcriptional difference between prokaryotic and eukaryotic cell
| Prokaryotic cell | Eukaryotic cell |
| A single type of RNA – polymerase synthesize all different type of RNA. | 3 different type of RNA – polymerase involved in simple type of RNA.
rRNA, mRNA and tRNA. |
| It take place within the cytoplasm. | It take place within the nucleus. |
| Transcription and Translation can be coupled. | Transcription and Translation cannot be coupled. |
| RNA do not undergo post transcriptional changes. | Post transcriptional changes takes place. |
| It is a simple process. | It is a complex process. |
| Require σ factor for initiation. | Required different transcription factor for initiation. |
Chromatin
There are two types of Chromatin-
| Euchromatin | Heterochromatin |
| Loosely bound DNA. | Tightly bound DNA. |
| Light strain. | Dark strain. |
| Demethylated and acetylated DNA. | Methylated and deacetylated DNA. |
| Transcriptionally active DNA. | Transcriptionally inactive DNA. |
| Replicated early in S phase. | Replicated late in S phase. |
Types of RNA
mRNA
known as messenger RNA.
It carries the message from DNA to ribosome for the synthesis of a protein.
It carries the information in the coded form.
mRNA is present in the least amount in a cell or least RNA of total RNA content in a cell.
Its 5’ contain unusual base pair and the 3’ Poly A tail present.
tRNA
known as transfer RNA.
It transfer the amino acid from cytoplasm to the ribosome.
It form 10 – 15% of total RNA content.
It is the smallest RNA among all the RNA and made up of only 90 – 95 basepair.
It contain some unusual base pair.
- Dihydrouracil (DHU)
- Pseudo uracil (ψ)
- Ribothymidine
- 1 – methyl guanosine
- Is pentyl Adenine
The structure of tRNA in 2D is clover leaf structure.
The structure in 3D is L – shape structure.
tRNA is known as Adapter molecule because from 1 end it lead the codon of the mRNA and on other end it bind the specific amino acid i.e. coded by that codon.
It is known as soluble RNA, as it is present in the soluble form in the cytoplasm.
rRNA
known as ribosomal RNA.
It form the ribosome.
It form 80 – 85% of total content.
16s rRNA recognise the 5’ of the mRNA and help in the initiation of process of translation.
5s rRNA form the tRNA binding site.
23s rRNA, it act as ribozyme and help in synthesis of peptide bond between the amino acid.
Genetic Code
Contribution of different scientist in the discovery of genetic code
George Gamow – He discovered genetic code are made up of 3 nucleotide, means they are triplet in nature.
Har Gobind Khurana – He develop chemical method for the synthesis of photopolymer and co – polymer RNA strand.
Nirenberg’s – He develop a cell free system for protein synthesis.
Severo Ochoa – He synthesize an enzyme known as polynucleotide phosphorylase that synthesize RNA with a defined sequence in a template independent manner.
Property of Genetic code :
- Genetic code are always triplet.
- They are always universal.
- A single amino acid may be coded by more than 1 codon. Hence, the code is degenerate.
- Genetic code are always in a continuous manner and no any punctuation present between the code.
- Out of the 64 codon, 61 are sense codon and 3 are non – sense codon.
- AUG is known as initiator codon and it has dual function in protein synthesis.
- Genetic code are unambiguous. It means a genetic code is specific for a kind of amino acid.
Mutation
There are two types in Gene mutation.
Point mutation –
In point mutation, a base pair is replace by another base pair whole frame of genetic code remains same but a single genetic code get change at the point of mutation.
Frame shift mutation –
In frame shift mutation, due to the addition or deletion of a base pair the whole frame of a genetic code get change beyond the point of mutation.
Genetic code are always triplet and if addition or deletion of base pair takes place in multiple of 3 in between the coding sequence of mRNA then, it does not lead to change in frame of mRNA.
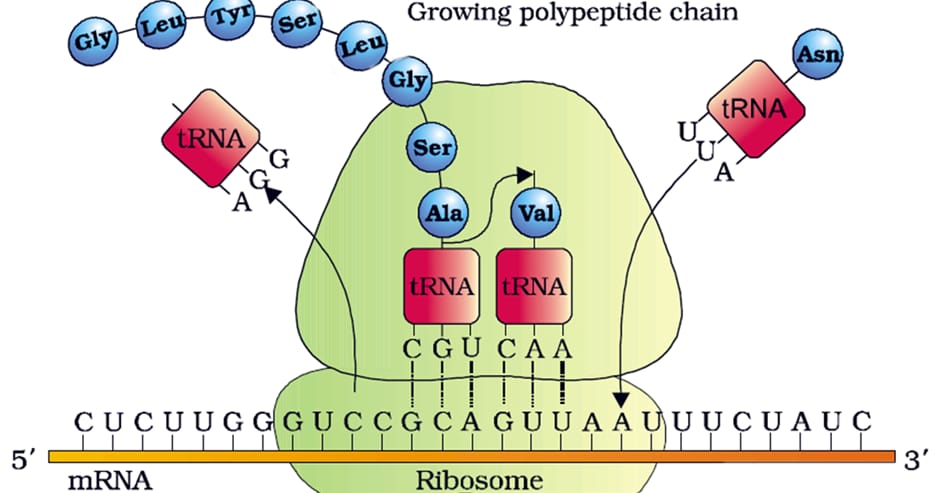
Translation
The process of formation of protein or conversion of a polynucleotide coded information into the polypeptide chain.
Aminoacylation of tRNA
It is also known as charging of tRNA or activation of tRNA.
The process of binding of a specific amino acid to its specific tRNA is known as aminoacylation of tRNA.
It carried out in two steps :
Activation of amino acid
AA + E + ATP → AA – AMP – E + Ppi
AA = amino acid, E = enzyme
AA – AMP – E = enzyme substrate complex
Charging of tRNA
AA – AMP – E + tRNA → tRNA – AA + AMP + E
Regulation of Gene expression
In eukaryotic cell, a gene expression is regulated at 4 different levels :-
- At the level of transcription.
- At the level of post – transcriptional change.
- At the level of transportation from nucleus to cytoplasm.
- At the level of Translation.
In prokaryotic cell, it is regulated in 2 ways :-
- At the level of transcription.
- At the level of translation.
Factors affecting gene regulation –
- Metabolism within body
- Environmental condition
- Physiology of organism
Operon – A cluster of gene that have a single operator, promoter and regulator gene.
Regulator gene is also known as inhibitor gene because when the gene express, it form repressor protein that inhibit the expression of structural gene.
Operator – Part of the transcription unit where repressor protein get bind.
It is present between promoter and structural gene.
Lac operon
A cluster of gene whose expression involved in lactose metabolism.
Lac operon is a inducible system and lactose itself act as a inducer.
Lac operon contains one regulatory gene and three structural genes (z, y and a).
i gene codes for the repressor of the lac operon
z gene codes for the β – galactosidase, that is responsible for the hydrolysis of the disaccharide, lactose into its monomer units, galactose and glucose.
The y gene codes for permease, which increases permeability of the cell to β – galactosidase.
The a gene codes for transacetylase.
The substrate for the enzyme β – galactosidase is Lactose and it regulates switching on and off of the operon. Here, lactose is termed as inducer.
In absence of carbon source like glucose, if lactose is provided in the growth medium of the bacteria, the lactose is transported to the cells through the action of permease.
From the i gene, repressor of the operon is synthesised.
To the operator region, repressor protein get bind and prevents RNA polymerase from transcribing the operon.
The repressor is inactivated by interaction with the inducer in the presence of an inducer, such as lactose or Allolactose.
It allows RNA polymerase access to the promoter and transcription proceeds
Regulation of lac operon is referred to as negative regulation.
Human Genome Project
It is a megaproject because –
- It take 13 years to get completed.
- The cost of the project is very high i.e. approximately.
Bioinformatics
HGP (Human genome project) give rise to the bioinformatics.
It is the branch of biology that store the biological information.
History of Human Genome Project
- HGP started in 1990 and initially led by Watson.
- In the beginning, HGP coordinated by USA department of energy and national institute of health then very soon coordinated by welcome trust of U.K.
- Later on other countries like Japan, Germany, France, China, India and other countries coordinate with the project.
Methodology
In the methodology, two different approach is used.
EST (Express sequence Tags)
In this approach, the DNA that express in the form of RNA is identified. The coding region (exon) of a gene is identified.
In this method, RNA from the cell cytoplasm is isolated and sequences.
Sequence Annotations
In this approach, whole DNA sequence identified and function of each region of DNA sequence is identified and attach with the sequence.
Steps of Sequence annotations-
- Whole DNA genome isolated from the cell.
- DNA is fragmented by the use of Restriction enzyme.
- DNA fragment inserted within the bacteria and the yeast cell by the use of BAC and YAC vector respectively for the gene cloning.
BAC – Bacterial artificial chromosome
YAC – Yeast artificial chromosome
- DNA fragment sequence by using automated DNA sequencer i.e. based on a principle develop by Frederick Sanger.
- Then, DNA sequence arranged on the basis of their overlapping region and is done by using specific computer program.
- Sequence are annotated and assigned to the chromosome they belong.
In May 2006, chromosome 1 is sequenced completely and announced.
In human, all 24 chromosome are sequenced.
(22 – autosome + X and Y chromosome).
Goals of Human Genome Project
- To find out the sequence of chemical base pair.
- To find out the total number of gene present in the Human Genome.
- To store the information in data base.
- To improve the data analysing programme.
- To improve the data transfer system to the different sectors.
- To address Ethical, Legal and Social issues (ELSI) that may arise during the human genome project.
Salient features of Human Genome Project
- Human genome is made up of 3164.7 million base pair.
- Human DNA have 30,000 total number of gene.
- In human genome, average gene size is 3000 bp.
Dystrophin gene i.e. present on the X – chromosome is the largest size gene and made up of 2.4 million base pair.
- Large part of the DNA is made up of repetitive DNA sequence.
- 50% DNA sequence function is unknown.
- Less than 2% DNA code for the protein.
- Chromosome number 1 has the highest no. of gene i.e. 2968 and chromosome Y has least no. of gene i.e. 231.
- In the Human genome, 1.4 million SNP (Single nucleotide polymorphism) and they are mainly pronounced as “Snips”.
DNA Fingerprinting
DNA sequence is divided into two parts:-
- Unique DNA – These are the DNA sequence that present once in the DNA.
- Repetitive DNA – These are the DNA sequence that repeat many times within the DNA.
It is of two types:-
- Interspread – It spreads within the whole DNA.
- Satellite DNA or Tandem repeat – Repetitive DNA sequence that are present adjacent to each other.
On the basis of length and composition of the repetitive DNA, there are two types :-
- Microsatellite – Length of the repeat is 1 – 6 bp.
It present many times within the DNA.
- Mini satellite – Length of the repeat is 11 – 60 bp.
Comparative less no. of repeat present in the DNA.
DNA polymorphism
Variation at a locus of DNA with 0.01 frequency in a population.
These variation are inherited and passes from parents to their off springs.
Mutation is one of the reason behind the DNA polymorphism.
In an individual, number of tandem repeat is unique and no two individual have the same no. of tandem repeats in all the chromosome except monozygotic twins.
During the HGP, it was identified that 99% DNA sequence is identical in all human beings and they vary from each other on the basis of 0.1% difference in chemical sequence of the DNA.
These variations are present within regulatory or non – coding region of the DNA.
These variations also present in the repetitive DNA and it give rise to VNTR.
Repetitive DNA is present within the centromere or telomeric region of the chromosome.
- Initially, DNA fingerprinting was develop by Alee Jeffreys.
DNA fingerprinting is based on southern hybridization and it is done by using VNTR probe.
DNA fingerprinting/DNA profile is the identification of the some specific region in the DNA sequence that is unique in an individual of the population.
Steps of DNA fingerprinting
- DNA isolation
- Fragmentation of the DNA by the use of R.E. enzyme.
- Separation of DNA fragment by gel electrophoresis.
- DNA fragment are transferred from Agarose gel to the nylon or nitrocellulose membrane and DNA is converted into single strand DNA.
- Hybridization is performed by the use of VNTR radioactive probe.
- Identification of hybridize DNA by passing UV rays and it is known as autoradiography.
Application of DNA fingerprinting
- It is use in the forensic lab in the murder and rape case.
- It is use in parental diagnosis.
# Molecular basis of Inheritance
# Molecular basis of Inheritance Class 12 Biology
# Molecular basis of Inheritance Notes
# Molecular basis of Inheritance NCERT Solutions
# Molecular basis of Inheritance Class 12
Do share this post if you liked Molecular basis of Inheritance. For more updates, keep logging on BrainyLads