- The transform ants growing on ampicillin containing medium are then transferred on a medium containing tetracycline.
- The recombinants will grow in ampicillin containing medium but not on that containing tetracycline.
The non-recombinants will grow on the medium containing both the antibiotics.
In this example, one antibiotic resistance gene helps in selecting the transformants whereas, the other antibiotic resistance gene gets ‘inactivated due to insertion’ of alien DNA and helps in selection of recombinants.
* Selection of recombinants due to inactivation of antibiotics is a cumbersome procedure, so alternative selectable markers are developed which differentiate recombinants from non-recombinants on the basis of their ability to produce colour in the presence of a chromogenic substrate.
-» In this method, a recombinant DNA is inserted within the coding sequence of an enzyme J3-galactosidase.
-» This results into inactivation of the enzyme, P-galactosidase (insertional inactivation).
-> The bacterial colonies whose plasmids do not have an insert, produce blue colour, but others do not produce any colour, when grown on a chromogenic substrate.
(c) Cloning sites are required to link the alien DNA with the vector.
• The vector requires very few or single recognition sites for the commonly used restriction enzymes.
• The presence of more than one recognition sites within the vector will generate several fragments leading to complication in gene cloning.
(d) Vectors for cloning genes in plants and animals are many which are used to clone genes in plants and animals.
• In plants, the Tumour inducing (Ti) plasmid of Agrobacterium tumefaciens is used as a cloning vector.
-» Agrobacterium tumefaciens is a pathogen of several dicot plants.
-» It delivers a piece of DNA known as T-DNA in the Ti plasmid which 1 transforms normal plant cells into tumour cells to produce chemicals
required by pathogens.
• Retrovirus, adenovirus, papillomavirus are also now used as cloning vectors in animals because of their ability to transform normal cells into cancerous cells.
Competent Host Organism (for transformation with recombinant DNA) is required because DNA being a hydrophilic molecule, cannot pass through cell membranes, Hence, the bacteria should be made competent to accept the DNA molecules,
(i) Competency is the ability of a cell to take up foreign DNA.
(ii) Methods to make a cell competent are as follow.
(a) Chemical method In this method, the cell is treated with a specific concentration of | a divalent cation such as calcium to increase pore size in cell wall.
The cells are then incubated with recombinant DNA on ice, followed by placing them briefly at 42°C and then putting it back on ice. This is called heat shock treatment.
• This enables the bacteria to take up the recombinant DNA.
(b) Physical methods In this method, a recombinant DNA is directly injected into the nucleus of an animal cell by microinjection method.
• In plants, cells are bombarded with high velocity microparticles of gold or tungsten coated with DNA called as biolistic or gene gun method.
(c) Disarmed pathogen vectors when allowed to infect the cell, transfer the recombinant
DNA into the host.
Different Methods to Introduce Alien/ Foreign DNA into Host Cells
There are three methods to introduce alien DNA into host cells namely;
- Micro Injection
- Biolistics or Gene Gun
- Disarmed Pathogen
Micro Injection-in this method the recombinant DNA is directly injected into the nucleus of an animal cell.
Biolistics (Gene Gun)- in this method, the cells are bombarded with high velocity micro- particles of gold or tungsten coated with DNA. This method is suitable for plants.
‘Disarmed Pathogen’ Vectors-when these vectors infect the cell, it transfers the recombinant DNA into the host.
What is Recombinant DNA technology?
Recombinant DNA technology is also known as Genetic Engineering. It is the process of joining together two DNA molecules from two different organisms. This is known as the recombinant DNA.
The steps involved in the processes of Recombinant DNA technology are:
- Isolation of DNA
- DNA fragmentation using restriction endonucleases
- Ligation of the desired DNA fragment into the vector
- Transfer of the recombinant DNA into the host
- Culture of the transformed cells in a nutrient medium.
- Extraction of the desired product.
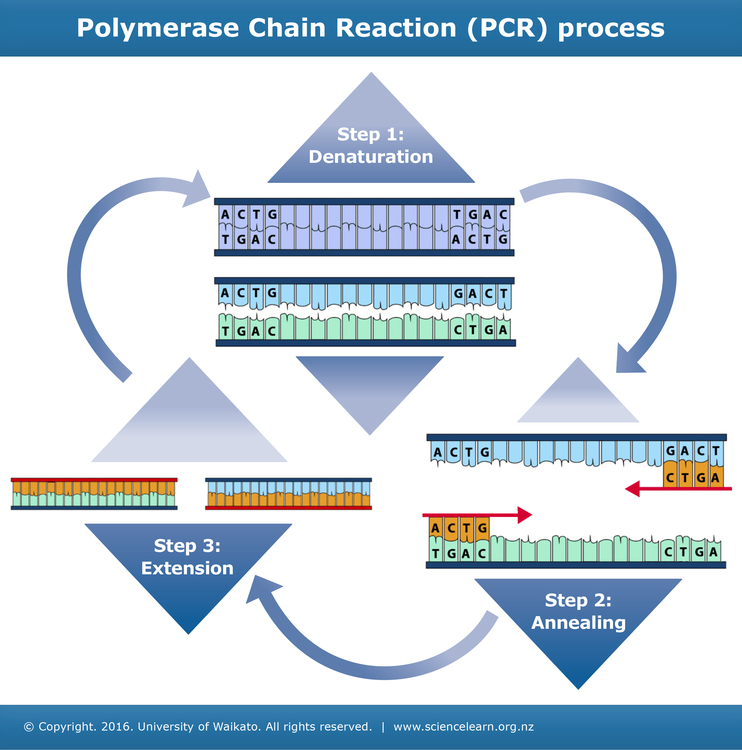
Steps in PCR technique
Step 1: Denaturation
As in DNA replication, the two strands in the DNA double helix need to be separated.
The separation happens by raising the temperature of the mixture, causing the hydrogen bonds between the complementary DNA strands to break. This process is called denaturation.
Step 2: Annealing
Primers bind to the target DNA sequences and initiate polymerisation. This can only occur once the temperature of the solution has been lowered. One primer binds to each strand.
Step 3: Extension
New strands of DNA are made using the original strands as templates. A DNA polymerase enzyme joins free DNA nucleotides together. This enzyme is often Taq polymerase, an enzyme originally isolated from a thermophilic bacteria called Thermus aquaticus. The order in which the free nucleotides are added is determined by the sequence of nucleotides in the original (template) DNA strand.
The result of one cycle of PCR is two double-stranded sequences of target DNA, each containing one newly made strand and one original strand.
The cycle is repeated many times (usually 20–30) as most processes using PCR need large quantities of DNA. It only takes 2–3 hours to get a billion or so copies.